Welcome to the Ernst lab!
We conduct research on Candida albicans, which is one of the most important human fungal pathogens. We explore molecular mechanisms, by which the pathogen increases its virulence in the human host. Signal transduction pathways leading to various cell morphologies, as well as events at the fungal cell surface that are crucial for the infection process are the focus of our research.
Candida albicans?
Candida albicans is the major human fungal pathogen, causing tenacious superficial mycoses, as well as life-threatening systemic mycoses. Superficial infections of the skin and mucosal layers are common, such as "soor" in the oral cavity and vaginal candidiasis, which most women acquire at least once in their lifetimes. Systemic infections, in which multiple internal organs are affected, are of increasing importance in immunocompromised patients. Life expectancy of patients with systemic candidiasis and mycoses in general is poor, since antifungal therapies are limited. But why does C. albicans cause disease, unlike related harmless yeasts such as baker´s yeast? What are the molecular mechanisms, which contribute to its virulence and allow it to colonize and persist in the human body?
An amazing variety of cell shapes
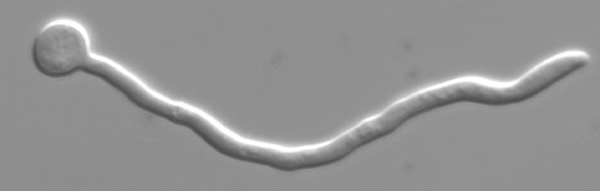
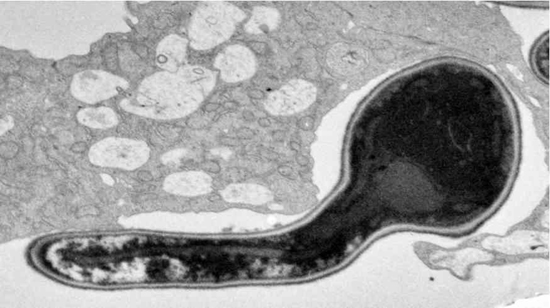
C. albicans has an amazing flexibility to alter its cellular forms and current data strongly suggest that this ability to change is of significant importance for its virulence. C. albicans is able to grow unicellularly in a yeast form and multiply by budding, very similar to baker´s yeast. In the presence of inducing compounds such as serum yeast cells rapidly develop true hyphae, which expand by continuous apical growth. In this growth form C. albicans resembles molds such as Aspergillus or Penicillium. Hyphal filaments allow C. albicans to firmly anchor in host cells and to penetrate endothelial and epithelial layers. In certain environments the pathogen grows as pseudohyphae, characterized by elongated yeast cells, which propagate by unipolar budding and remain attached, thus giving this growth form a "beads on a chain"-appearance. Some isolates of C. albicans are able to spontaneously switch between a "white", typical yeast-cell form, and an "opaque"-form, which appears as short rods. The reasons and functions of this "phenotypic switching" are intensely investigated at the present time. Finally, in certain environments C. albicans is able to form thick-walled chlamydospores, which are of yet unknown importance for its biology and virulence.
Signalling pathways
How are external environmental cues translated into the cellular program of differentiation that models a yeast form into a hypha? Current views state that several signalling pathways act in concert to trigger this morphological switch. Hypha induction by serum or certain sugars present in host cell membranes, such as N-acetylglucosamine, critically requires a protein kinase A (PKA) signalling pathway. A key regulator of the several morphological forms of C. albicans, Efg1, was isolated by our group. Efg1, is activated by protein kinase A and sets the stage for the induction of hypha-specific genes, while repressing yeast-specific genes. In addition, a "mitogen-activated protein" (MAP)-kinase pathway also contributes to the development of true hyphae. Since hypha formation occurs at body temperature, at low cell density and at neutral pH, pathways must exist that convey these external signals. Which are the targets of these signalling pathways and how do they operate to allow the reprogramming of cellular activities in a sequential manner? In this search and analyses we take advantage of the knowledge of the complete C. albicans genomic sequence and the availability of DNA arrays containing all C. albicans genes (about 6000).
Cell surface events
The structure of the fungal cell surface determines the processes occurring during the contact with human host cells, such as adhesion, phagocytosis and penetration. We are in the process of analysing the PMT gene family of C. albicans, which encodes mannosyltransferases modifying secreted proteins by short mannose chains at serine or threonine residues (O-glycosylation). The PMT family encodes 5 Pmt isoforms, which appear to have different target specificities. Interestingly, the lack of the Pmt1-isoform not only blocks adhesion and abolishes virulence, but also leads to defective hypha morphogenesis and supersensitivity to antifungal drugs. But which protein targets are modified by specific Pmt proteins? One of the targets is the Msb2 membrane protein, which acts as a sensor to signal defects in glycostructures to the Cek1 MAP kinase, which in turn triggers “rescue” responses. Interestingly, the large external glycodomain of Msb2 is continuously shed into the environment and it can act to bind and thereby inactivate so-called antimicrobial peptides (AMP). AMPs are produced by the human host and serve as protectants against microbial invaders. It appears that C. albicans has found a way to deflect host responses by shedding the Msb2 glycofragment.
Applied aspects: C. albicans and C. utilis
Much of the recent interest in C. albicans is due to the need for effective new antifungal compounds. Our basic research on virulence traits of C. albicans reveals reactions necessary for fungal pathogenicity and viability; such reactions may be considered potential targets for novel antifungal compounds. In some cases, homologous components are missing in human cells; therefore, inhibition by a potential antifungal compound should be very selective to only attack the fungal invader and not cause side-effects in humans.
Although the name “Candida” is associated most with the fungal pathogen C. albicans it should not be overlooked that some Candida species are completely harmless and are even useful for biotechnological applications. For example, Candida utilis (also known as Torula yeast) is classified as a “GRAS” (generally recognized as safe) organism by regulatory authorities and it has already been used as a food additive and to produce several molecules and proteins (including recombinant proteins) from cheap, biomass-derived waste substrates. In our group we are in the process of further optimizing this Candida species for its use in heterologous protein production as an alternative to existing microbial production systems.
Funding
Our current research is funded by the following grants:
- ERA-NET Infect "FunComPath"
- MOI Graduate School "Molecules of Infection"
- Deutsche Forschungsgemeinschaft Er 47/13-1